Chapter 1 Introduction: Work Flow and Reproducible Analyses
Contributors: Ben Arnold
This handbook collates a number of tips to help organize the workflow of epidemiologic data analyses. There are probably a dozen good ways to organize a workflow for reproducible research. This document includes recommendations that arise from our own team’s experience through numerous field trials and observational data analyses. The recommendations will not work for everybody or for all applications. But, they work well for most of us most of the time, else we wouldn’t put in the time to share them.
Start with two organizing concepts:
Workflow. Defined here as the process required to draw scientific inference from data collected in the field or lab. I.e., the process by which we take data, and then process it, share it internally, analyze it, and communicate results to the scientific community.
Reproducible research. A fundamental characteristic of the scientific method is that study findings can be reproduced beyond the original investigators. Data analyses that contribute to scientific research should be described and organized in a way that they could be reproduced by an independent person or research group. A data analysis that is not reproducible violates a core principle of the scientific method.
1.1 Workflow
Broadly speaking, a typical scientific data science work flow involves four steps to transform raw data (e.g., from the field) into summaries that communicate results to the scientific community.
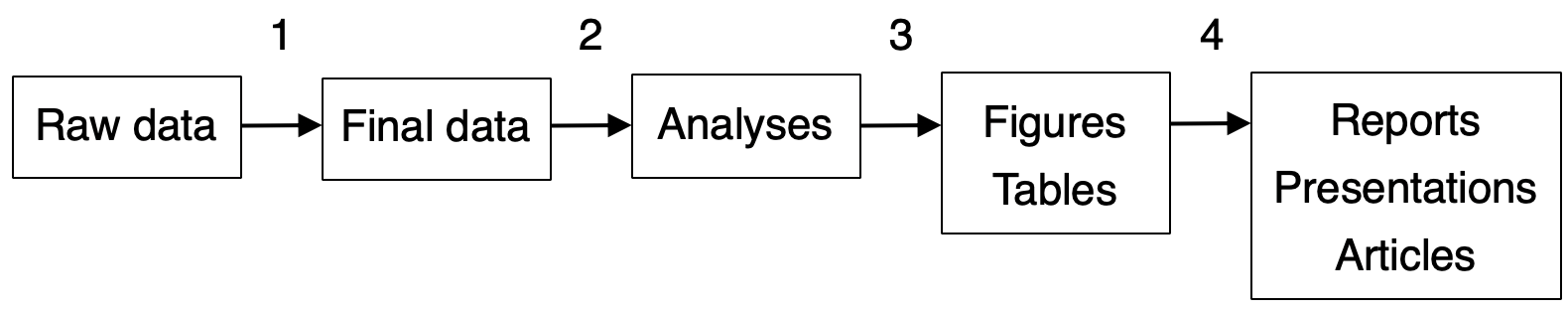
Figure 1.1: Overview of the four main steps in a typical data science workflow
When starting a new project, the work flow tends to evolve gradually and by iteration. Data cleaning, data processing, exploratory analyses, back to data cleaning, and so forth. If the work takes place in an unstructured environment with no system to organize files and work flow, it rapidly devolves into into a disorganized mess; analyses become difficult or impossible to replicate and they are anything but scientific. Projects with short deadlines (e.g., proposals, conference abstract submissions, article revisions) are particularly vulnerable to this type of organizational entropy. Putting together a directory and workflow plan from the start helps keep files organized and prevent disorder. Modifications are inevitable – as long as the system is organized, modifications are usually no problem.
Depending on the project, each step involves a different amount of work. Step 1 is by far the most time consuming, and often the most error-prone. We devote an entire chapter to it below (Data wrangling)
1.2 Reproducibility
As a guiding directive, this process should be reproducible. If you are not familiar with the concept of reproducible research, start with this manifesto (Munafo et al. 2017). For a deeper dive, we highly recommend the recent book from Christensen, Freese, and Miguel (2019). Although it is framed around social science, the ideas apply generally.
Essential reading: Ten Simple Rules for Reproducible Computational Research Please read this excellent paper on computational reproducibility from Sandve et al 2013. It encapsulates many of the practices described herein (we elaborate more, and provide concrete examples).
A key tool to help ensure your R code runs reliably on different machines and over time is renv (renv).
1.3 Automation
We recommend that the workflow be as automated as possible using a programming language. Automating the workflow in a programming language, and essentially reducing it to text, is advantageous because it makes the process transparent, well documented, easily modified, and amenable to version control; these characteristics lend themselves to reproducible research.
At Proctor, we mostly use R. With the development of Rstudio, R Markdown and the tidyverse ecosystem (among others), the R language has evolved as much in the past few years as in all previous decades since its inception. This has made the conduct of automated, reproducible research considerably easier than it was 10 years ago.
If you have a step in your analysis workflow that involves point-and-click or copy/paste, then STOP, and ask yourself (and your team): How can I automate this?